|
|
|
DEVELOPMENTAL BIOLOGY 3230 |
|
|
|
|
|
 |
|
 |
|
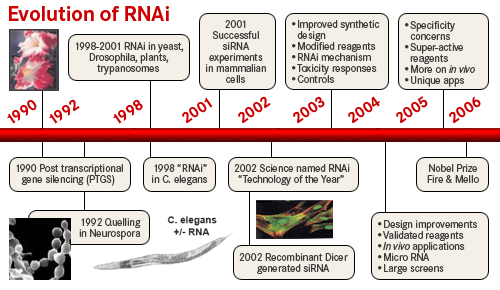
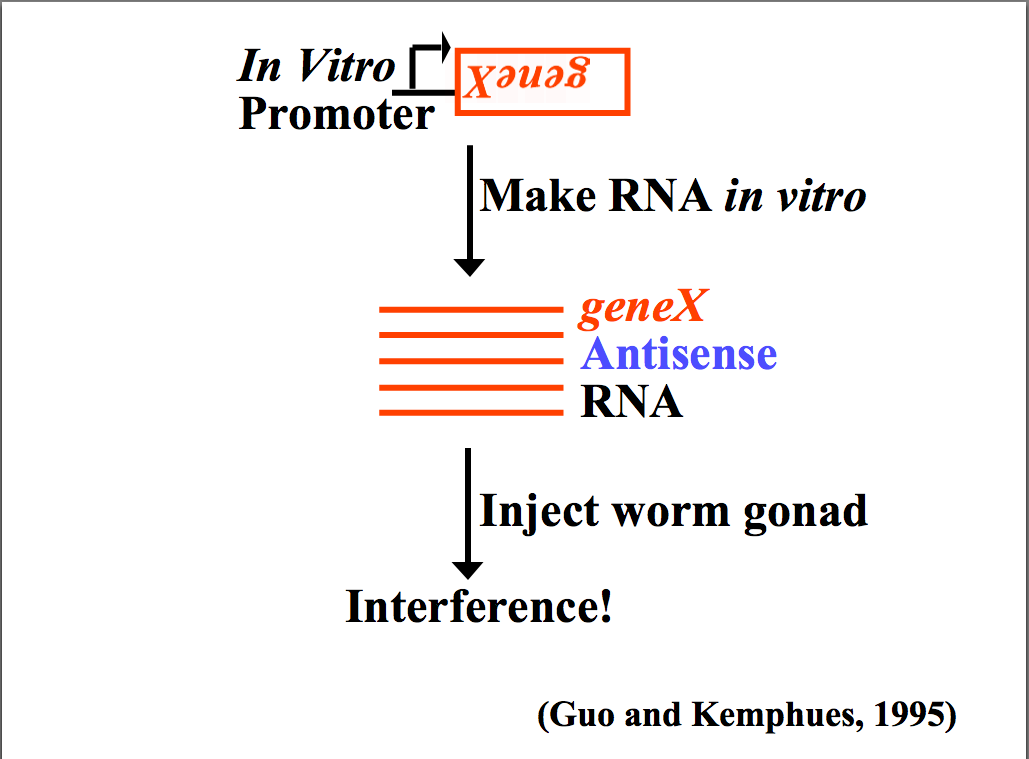
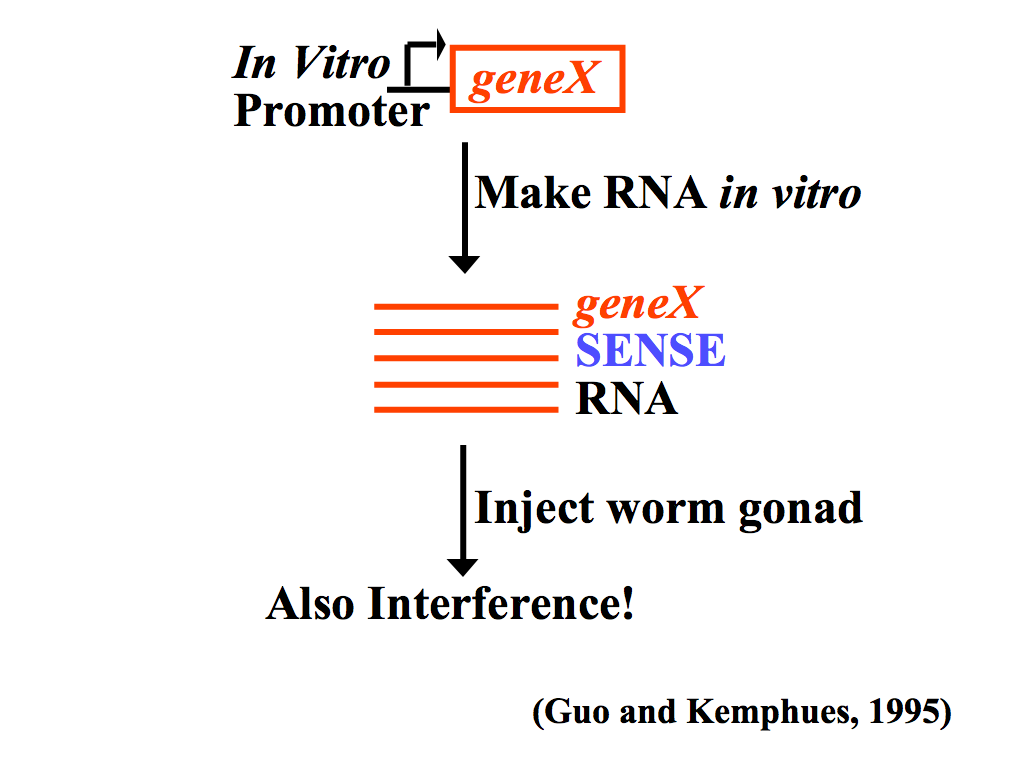
| In 1990 Napoli and Jorgensen got an unexpected result when they tried to "overexpress" a plant transgene, chalcone synthase (CHS), to verify that it was the rate limiting enzyme in anthocyanin biosynthesis that gives petunias their deep violet color. To their surprise the transgene caused "white" petunias! They found that the transgene "suppressed' both the injected and endogenous CHS to levels 50 fold lower than in "wild-type" petunias. They termed this phenonmenom "cosuppressing". A similar finding was reported in Neurospora crassa in 1992 when injected homologous RNA caused "quelling" of the endogenous gene. It was first described in animals (C. elegans) in 1995 when Guo and Kemphues found the surprising result that injection of either sense or antisense RNA to par-1 caused degradation of the endogenous par-1 mRNA. Up to this point antisense suppression was thought to function by binding to complementary mRNA sequences and targeting them for degradation. However, the "control" experiment of injecting "sense" RNA had the same effect!!
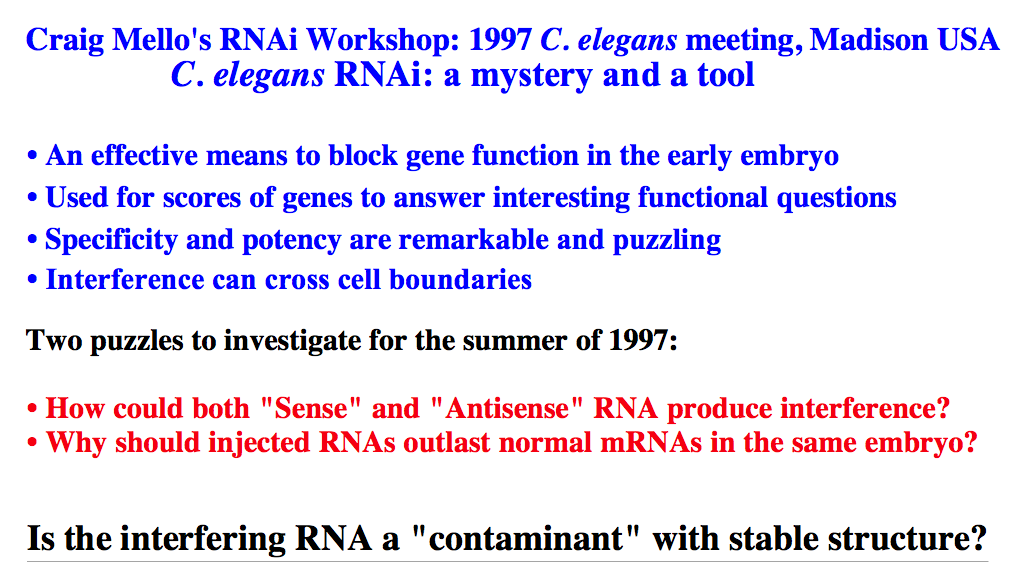
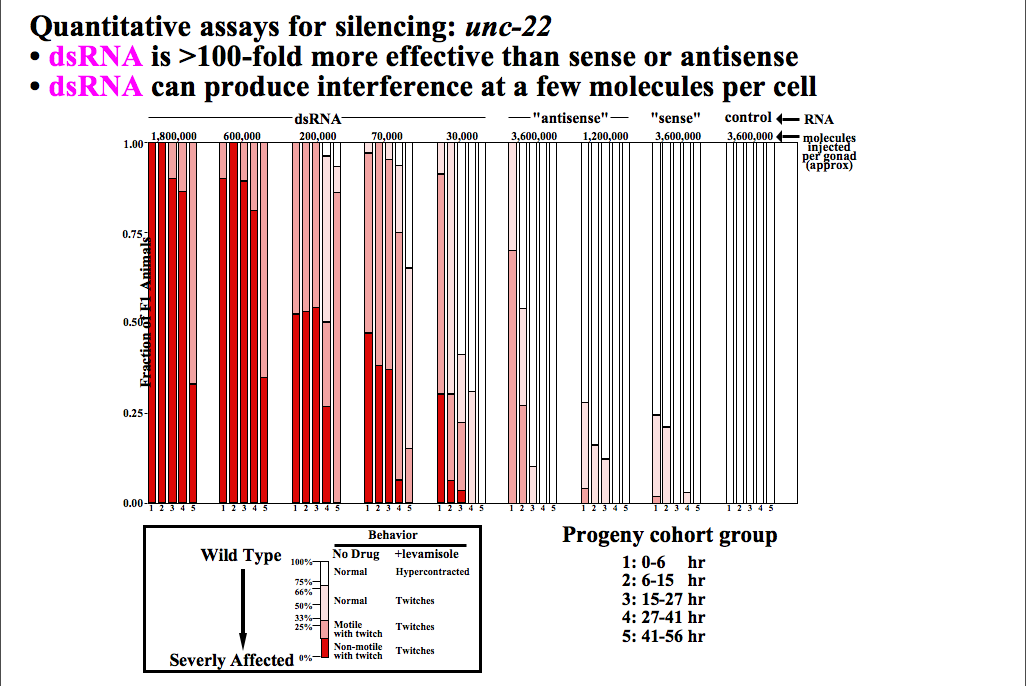
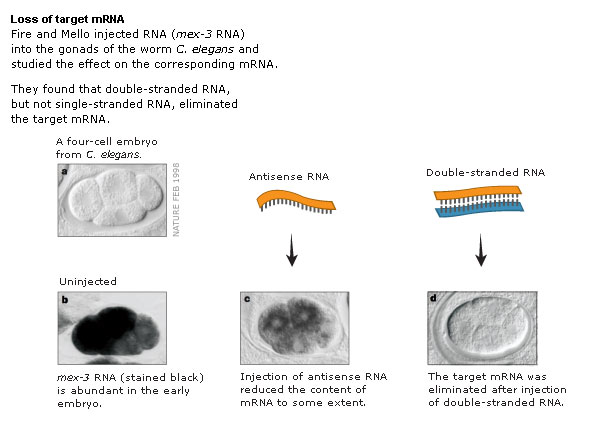
Fire and Mello in 1998 showed that the "real" silencing effector was dsRNA! They reasoned that in each of the earlier experiments there was the possibility of generating dsRNA contanminants for technical reasons. They designed experiments to carefully separated the effects of sense, antisense, and dsRNA in their "quelling" assay. They showed conclusively that it was in fact dsRNA that was the real trigger for "gene silencing". The dsRNA somehow caused the accelerated degradation of the endogenous mRNA, AND was somehow catalytic, AND could be inherited!! The field of RNA interference (RNAi) has exploded as a new powerful technique for experimentally regulating gene expression, exploring gene function, and as a basis for novel disease therapies.
|
|
|
| See the Noble site for summary of discovery, Noble lectures, and interviews |
|
| Nature RNAi Review Site and great RNAi animation (or local) |
|
|
|
The following 6 figures are from the Noble site illustrated presentation |
|
|
|
 |
|
|
|
|
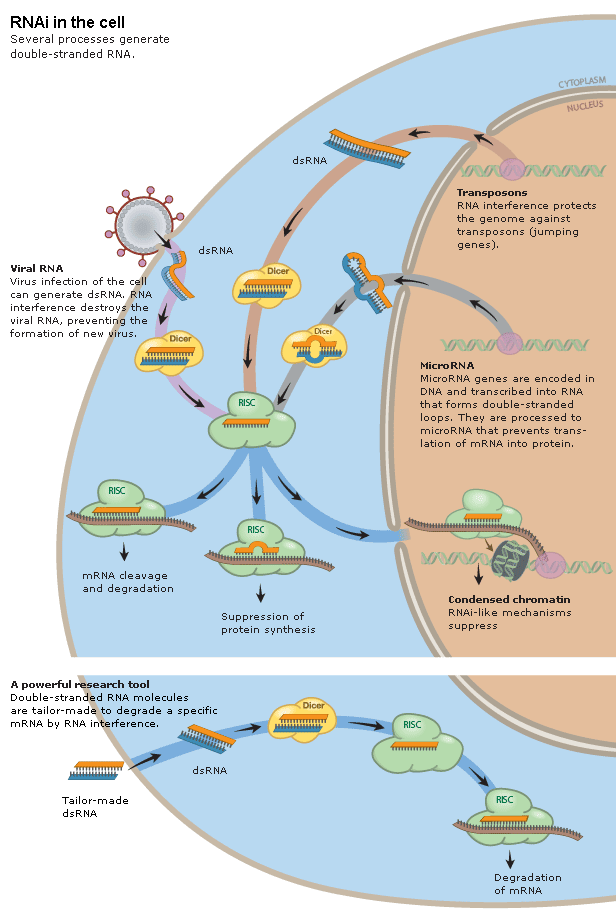 |
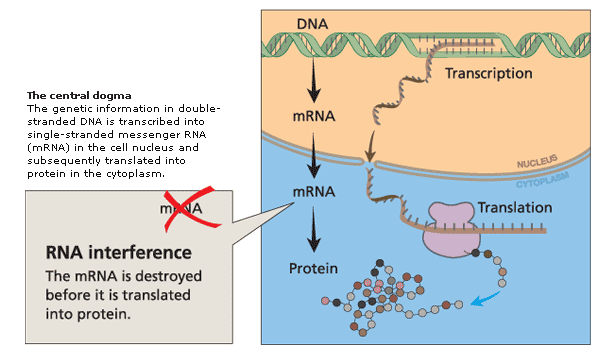
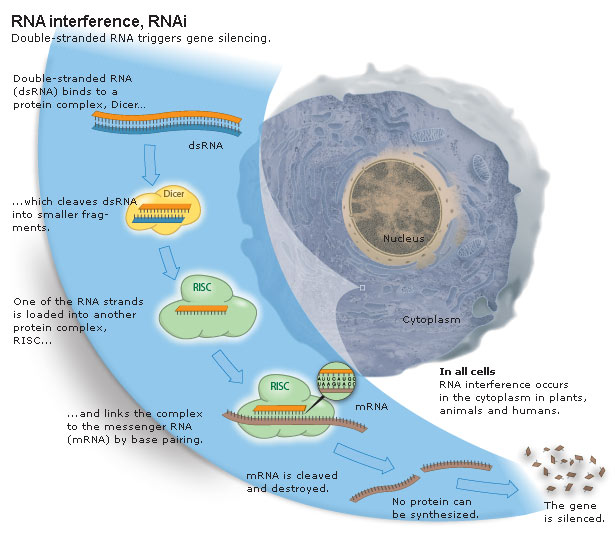
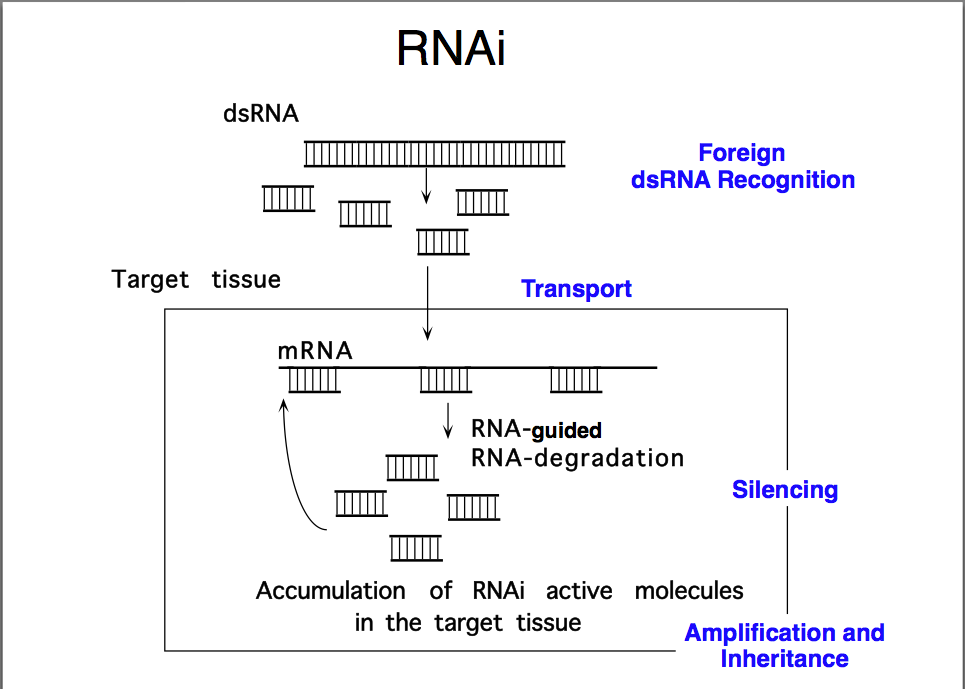
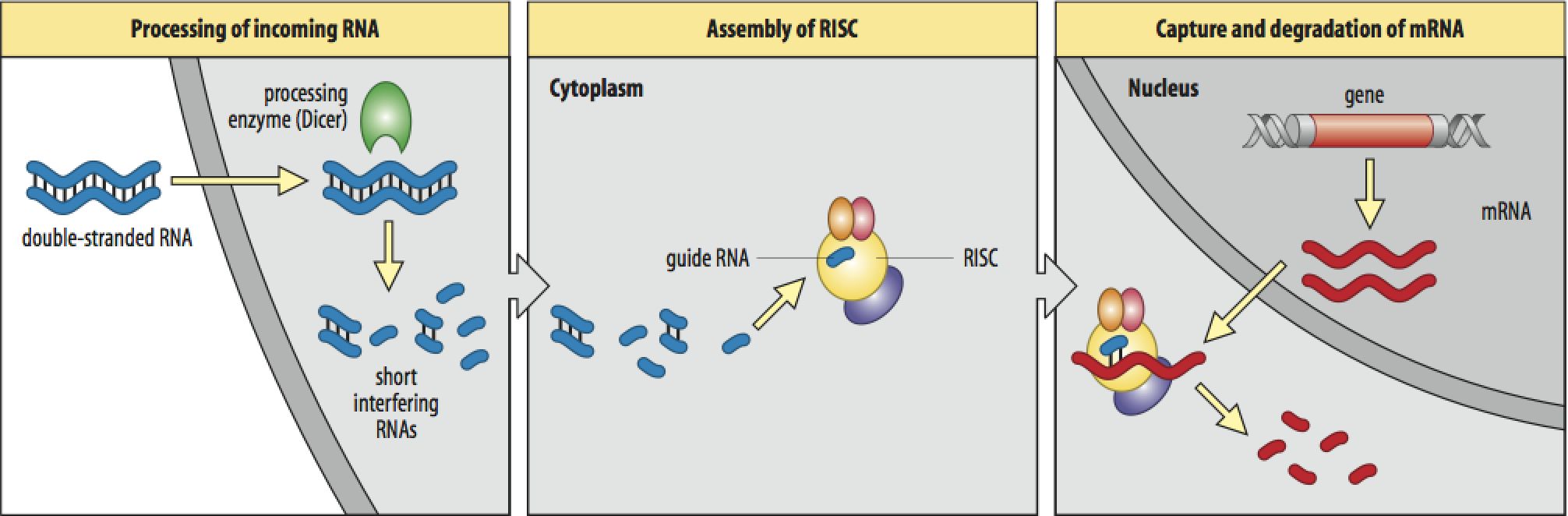
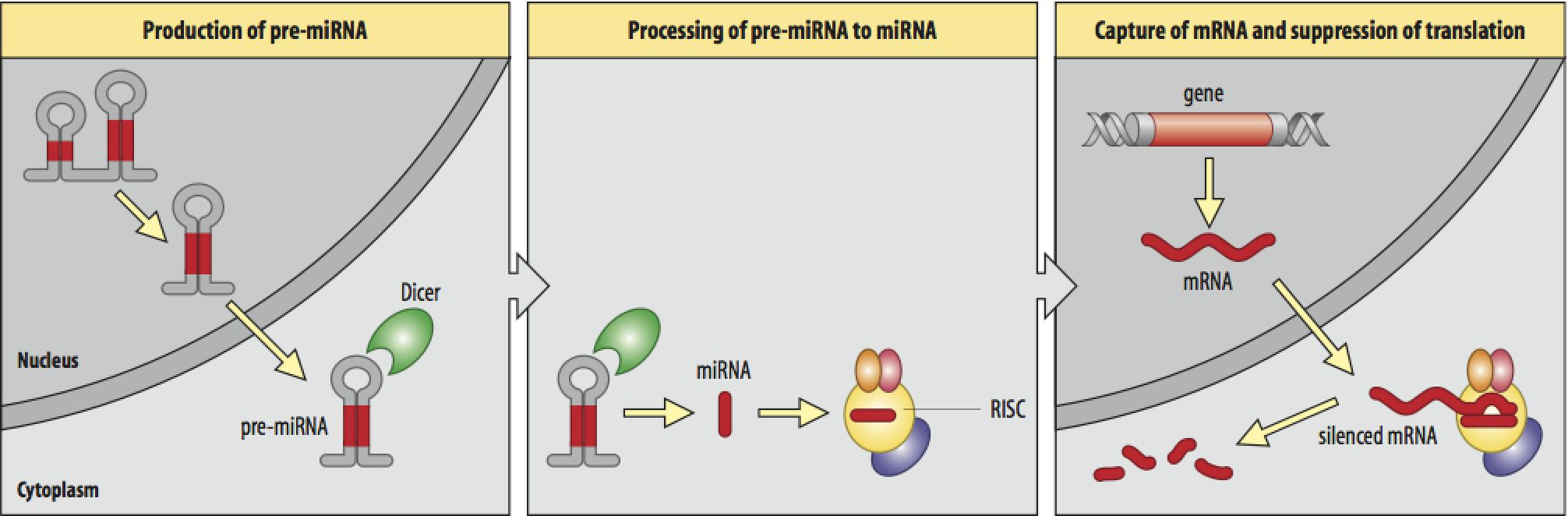
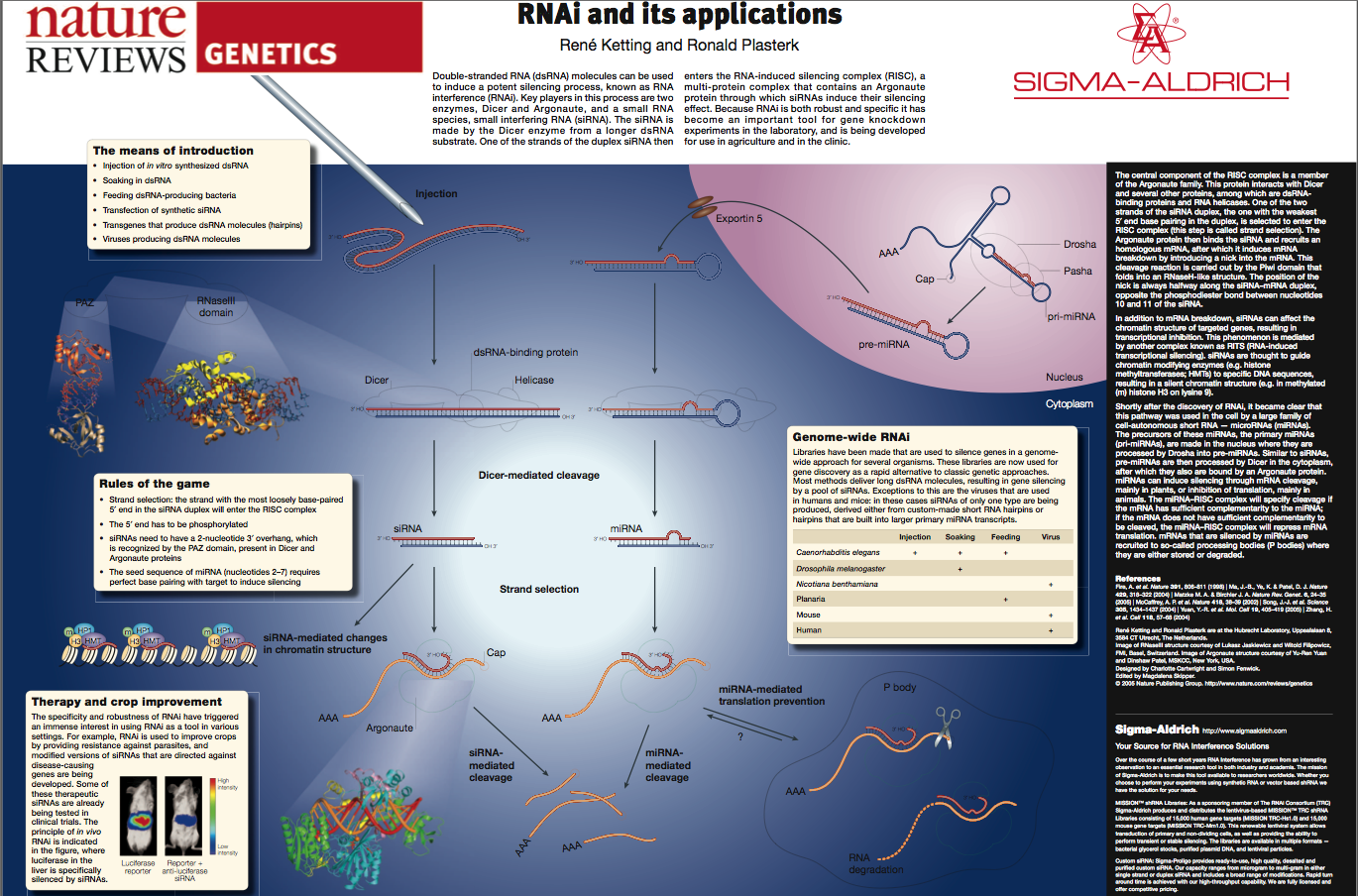
dsRNA is the key, but long dsRNA needs to be recognized by the Type III RNAse (dsRNA endonuclease) called "Dicer". Dicer recognizes long dsRNA and cleaves them into short 21-23 nt "siRNAs" (short interfering). Next the siRNA are loaded into the RISC complex (RNA Induced Silencing Complex) and a RNAase H enzyme called "Slicer" removes the sense strand. RISC now scans mRNAs in the P-body (mRNA decay center of cell) for complementary sequences. Binding of RISC targets the mRNA for rapid degradation. Single stranded mRNA fragments are recognized by a cytoplasmic RNA polymerase an generate more dsRNA that can be new targets for Dicer. In this way a dsRNA signal can be amplified and perhaps "inherited". The mechanism for cell to cell spreading is well documented, but not yet completely understood.
RNAi acts as a defense against viruses, transposon hopping, and part of the molecular machinery is also involved in mediating the function of microRNA genes. Over 1% of the genes in the mammmalilan genome are now thought to be encode microRNAs that regulate gene expression in part via the RNAi pathway.
|
|
| RNAi offers a new tool for influencing gene expression in cells where other genetic techniques are limited or unwieldly. RNAi can be used to easily look at the post embryonic and cell specific effects of gene "KO". Amazingly, feeding worms bacteria that express high levels of dsRNA can systemically suppress gene expression thoughout the worm cells! There are now "feeding" libraries that include bacterial clones for virtually every gene in the worm genome!! |
|
|
|
 |
|
|
|
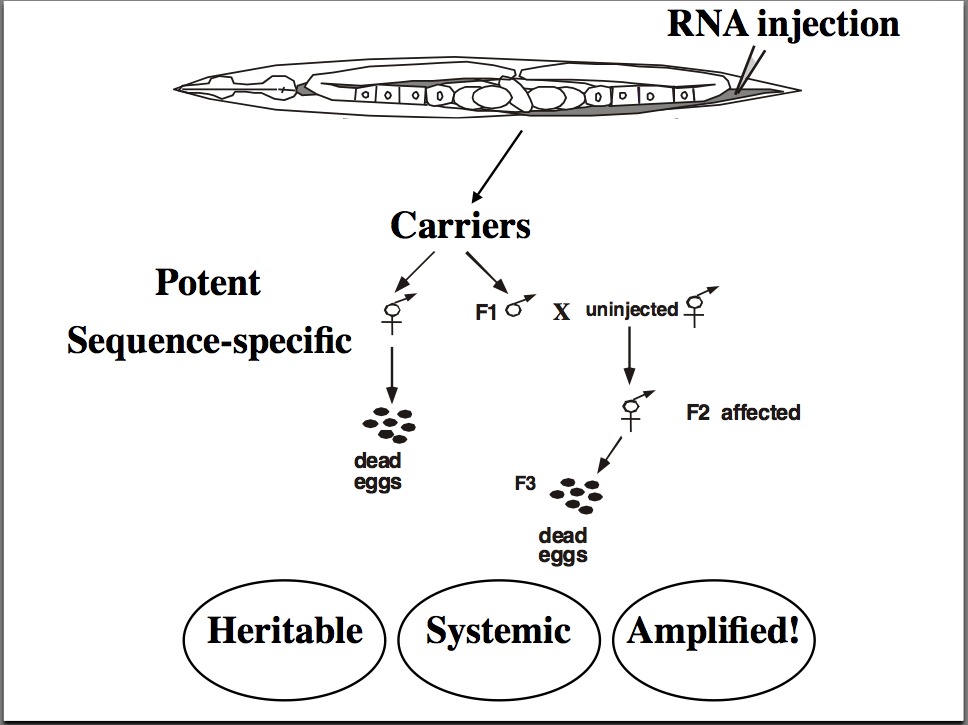
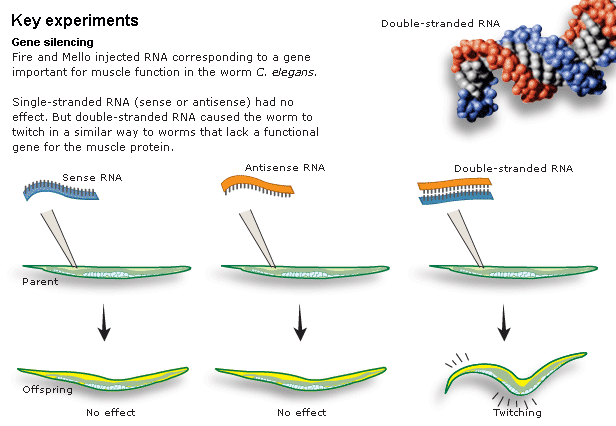
RNA injection into worm syncial gonad cause an amplified, systemic, and heritable suppression of the "cognate" gene. Thought to work via "antisense" RNA binding to mRNA and targeting it for degradation. But that doesn't explain the amplification, spread, and heritability of the effect! |
|
 |
|
|
|
Confusing results seen by Guo and Kemphues. Both the "experimental" antisense RNA and the "control" sense RNA caused gene suppression! How could this be if the mechanism is antisense RNA binding to the complementary mRNA and targeting it for degradation? |
|
 |
|
 |
|
|
|
 |
|
|
|
 |
|
|
 |
|
|
 |
|
|
 |
dsRNA is the trigger, but what are the other molecules that are involved? Must be RNAses specific for dsRNA. How do you find them? Several approaches were taken, including a cell/biochemical approach and a forward genetic screen approach. Both these avenues have lead to the identification of the molecules involved in the RNAi pathway. Dicer, Slicer, RISC, an others involved in the amplification, systemic transmission, and heritibility of RNAi. |
|
|
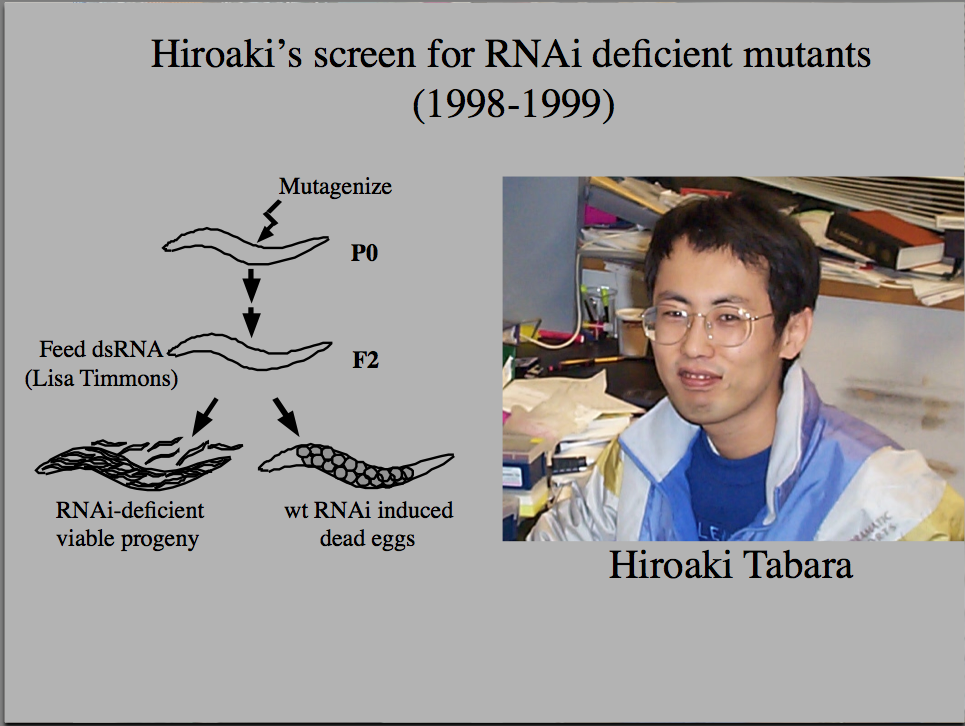
Tabara (in Mello's lab) used a forward genetic screen to identify genes needed for RNAi. He identified many genes, including Argonaute, that is now thought to be the "Slicer" component of RISC. |
|
|
|
|
 |
|
|
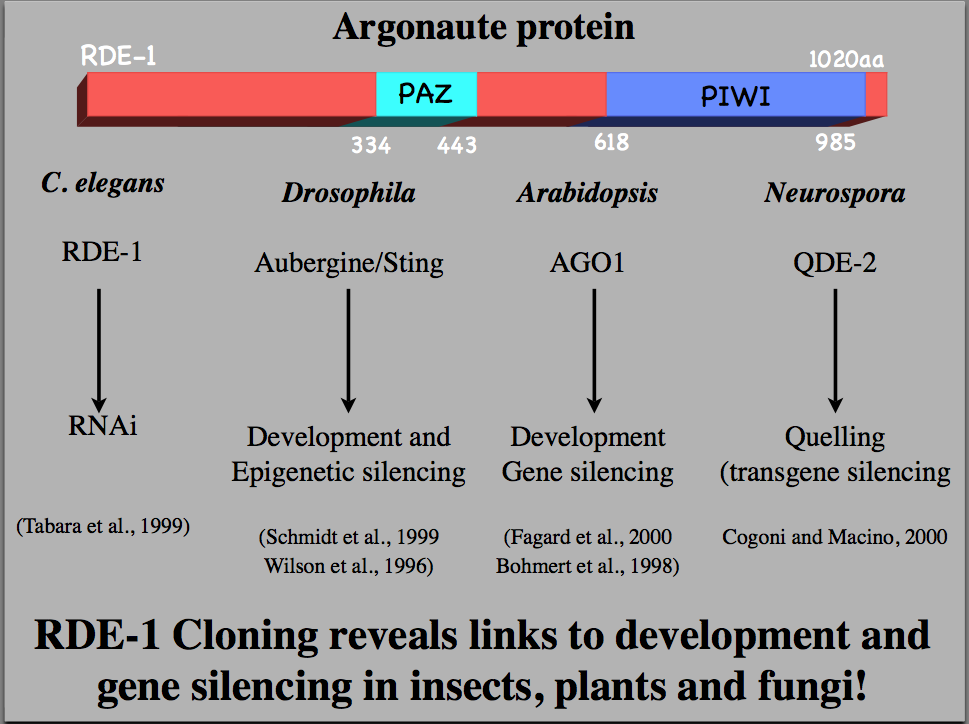 |
The conserved nature of Argonaute proteins involved in gene silencing during development and RNAi suggests a high degree of conservation of the RNAi mechanism among plants animals and fungi. |
|
|
|
 |
|
|
 |
|
 |
|
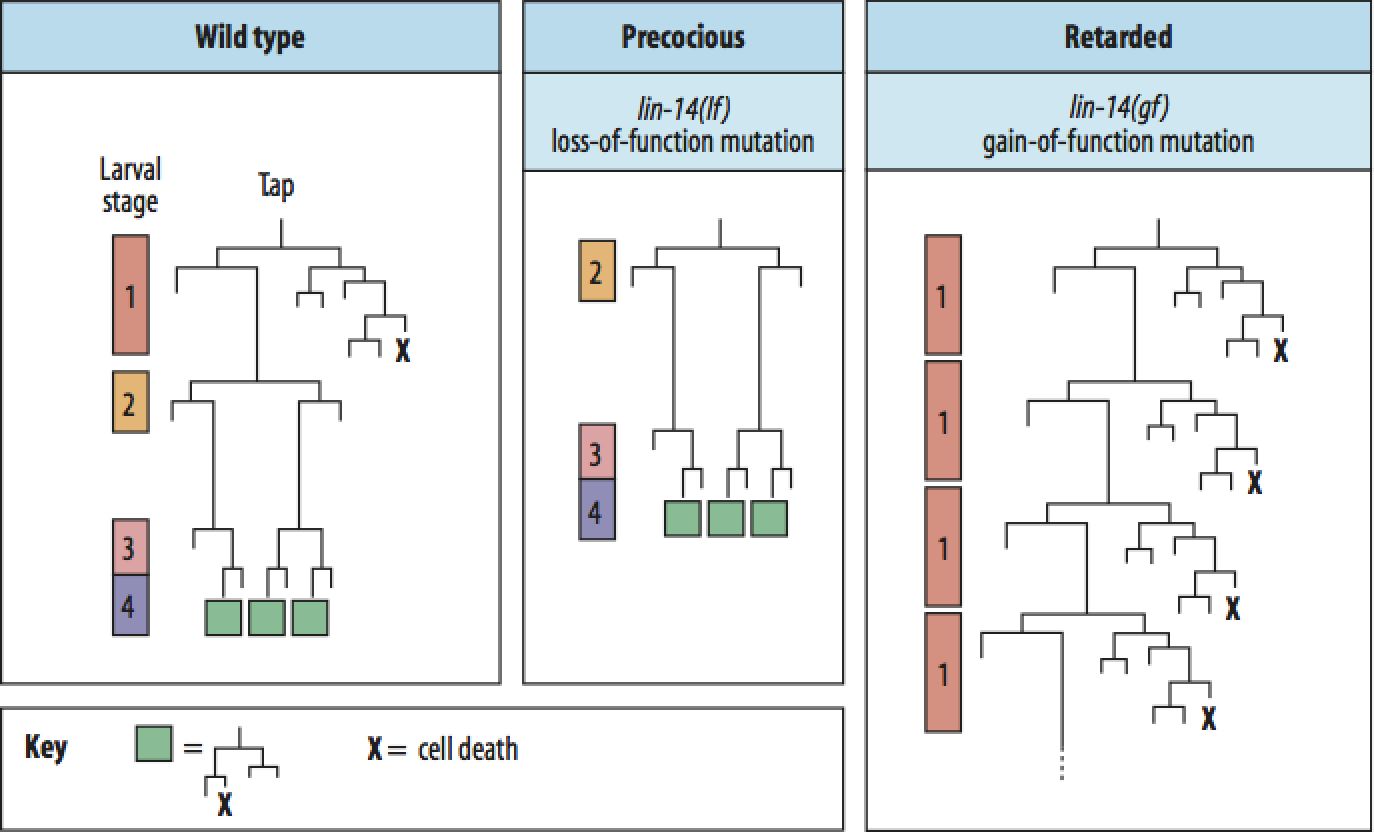 |
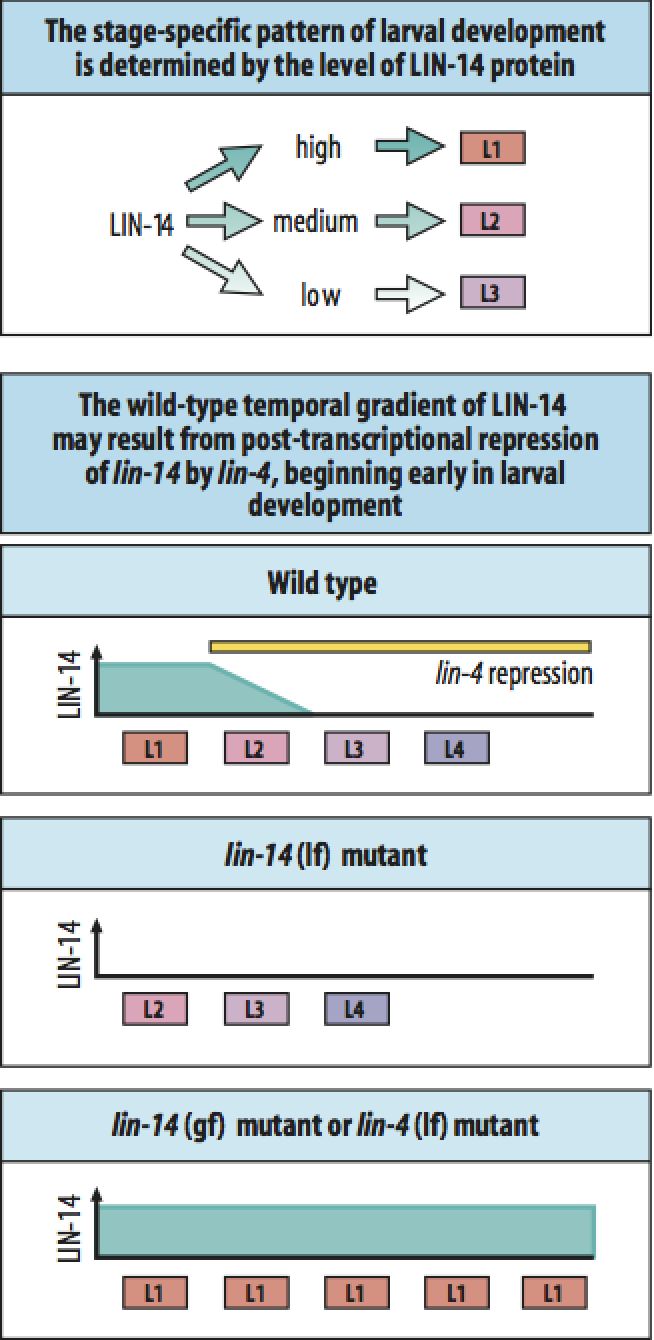
The first genes encoding microRNAs were discovered in C. elegans. lin-4 encodes a small microRNA that regulates lin-14. Mutations in these genes causes a "heterochronic" phenotype where lineages are "displaced" in time, either forward or backward. The timing of lineages is obviously important for the production of the correct cells at the correct time during development. |
|
|
 |
|
|
 |
|
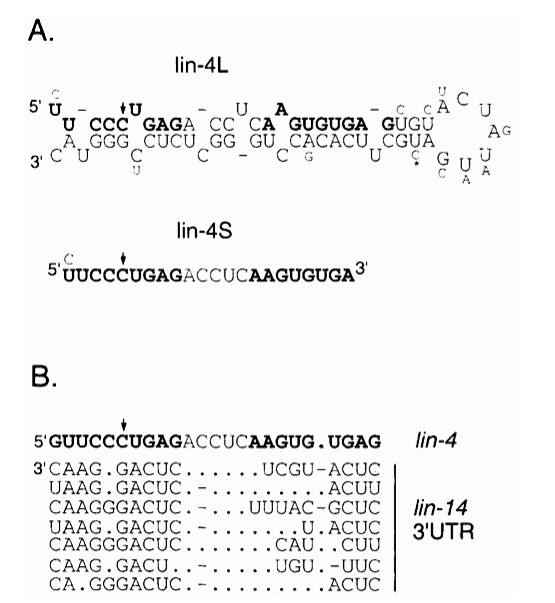
| The finding that lin-4 did not encode a protein, but a small dsRNA let to a model of RNA genes that regulate mRNA stability and translation. Evidence: no conserved ATG and stop. 3rd positions conserved (suggest RNA sequence and not AA sequence is conserved across species). Sequence is complementary to 3' UTR of lin-14 mRNA. This 3' UTR sequence is essential for lin-4 function and highly conserved across species! |
|
| Now estimated that over 1% of human genes encode miRNAs involved in gene regulation.
If there are so many genes encoding microRNAs why have they been so hard to discover?
TARGET SIZE!! Sequcence Specificity? Does it require multiple hits to prevent sufficient complementary? May require clever "genomics and infomatics" and use of RNAi defective mutants to identifiy all the microRNA genes. Just beginning to understand the importance of microRNA genes in animal development and cell biology.
|
|
 |
|
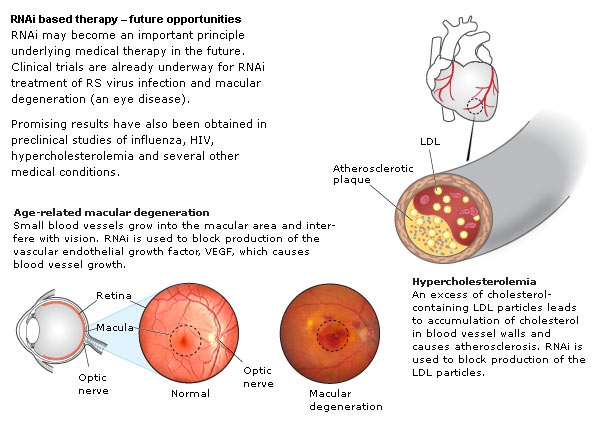
| The original link to this poster is at Nature Reviews RNAi |
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|





















